Adaline: Adaptive Linear Neuron Classifier
An implementation of the ADAptive LInear NEuron, Adaline, for binary classification tasks.
from mlxtend.classifier import Adaline
Overview
An illustration of the ADAptive LInear NEuron (Adaline) -- a single-layer artificial linear neuron with a threshold unit:
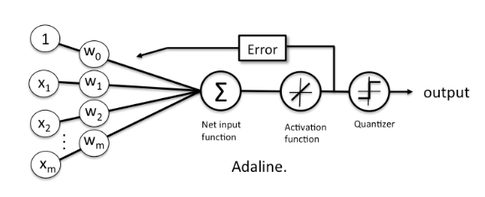
The Adaline classifier is closely related to the Ordinary Least Squares (OLS) Linear Regression algorithm; in OLS regression we find the line (or hyperplane) that minimizes the vertical offsets. Or in other words, we define the best-fitting line as the line that minimizes the sum of squared errors (SSE) or mean squared error (MSE) between our target variable (y) and our predicted output over all samples in our dataset of size .
LinearRegression implements a linear regression model for performing ordinary least squares regression, and in Adaline, we add a threshold function to convert the continuous outcome to a categorical class label:
$$y = g({z}) = $$
An Adaline model can be trained by one of the following three approaches:
- Normal Equations
- Gradient Descent
- Stochastic Gradient Descent
Normal Equations (closed-form solution)
The closed-form solution should be preferred for "smaller" datasets where calculating (a "costly") matrix inverse is not a concern. For very large datasets, or datasets where the inverse of may not exist (the matrix is non-invertible or singular, e.g., in case of perfect multicollinearity), the gradient descent or stochastic gradient descent approaches are to be preferred.
The linear function (linear regression model) is defined as:
where is the response variable, is an -dimensional sample vector, and is the weight vector (vector of coefficients). Note that represents the y-axis intercept of the model and therefore .
Using the closed-form solution (normal equation), we compute the weights of the model as follows:
Gradient Descent (GD) and Stochastic Gradient Descent (SGD)
In the current implementation, the Adaline model is learned via Gradient Descent or Stochastic Gradient Descent.
See Gradient Descent and Stochastic Gradient Descent and Deriving the Gradient Descent Rule for Linear Regression and Adaline for details.
Random shuffling is implemented as:
- for one or more epochs
- randomly shuffle samples in the training set
- for training sample i
- compute gradients and perform weight updates
- for training sample i
- randomly shuffle samples in the training set
References
- B. Widrow, M. E. Hoff, et al. Adaptive switching circuits. 1960.
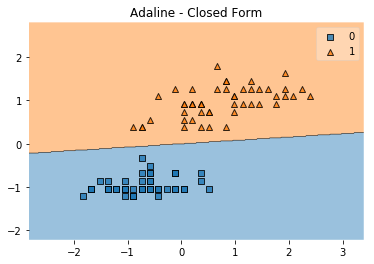
Example 1 - Closed Form Solution
from mlxtend.data import iris_data
from mlxtend.plotting import plot_decision_regions
from mlxtend.classifier import Adaline
import matplotlib.pyplot as plt
# Loading Data
X, y = iris_data()
X = X[:, [0, 3]] # sepal length and petal width
X = X[0:100] # class 0 and class 1
y = y[0:100] # class 0 and class 1
# standardize
X[:,0] = (X[:,0] - X[:,0].mean()) / X[:,0].std()
X[:,1] = (X[:,1] - X[:,1].mean()) / X[:,1].std()
ada = Adaline(epochs=30,
eta=0.01,
minibatches=None,
random_seed=1)
ada.fit(X, y)
plot_decision_regions(X, y, clf=ada)
plt.title('Adaline - Closed Form')
plt.show()
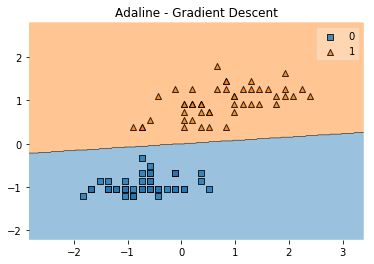
Example 2 - Gradient Descent
from mlxtend.data import iris_data
from mlxtend.plotting import plot_decision_regions
from mlxtend.classifier import Adaline
import matplotlib.pyplot as plt
# Loading Data
X, y = iris_data()
X = X[:, [0, 3]] # sepal length and petal width
X = X[0:100] # class 0 and class 1
y = y[0:100] # class 0 and class 1
# standardize
X[:,0] = (X[:,0] - X[:,0].mean()) / X[:,0].std()
X[:,1] = (X[:,1] - X[:,1].mean()) / X[:,1].std()
ada = Adaline(epochs=30,
eta=0.01,
minibatches=1, # for Gradient Descent Learning
random_seed=1,
print_progress=3)
ada.fit(X, y)
plot_decision_regions(X, y, clf=ada)
plt.title('Adaline - Gradient Descent')
plt.show()
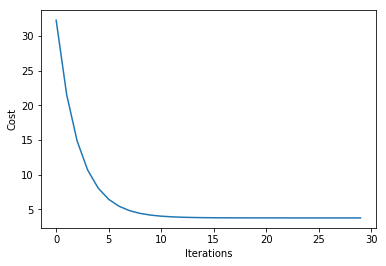
plt.plot(range(len(ada.cost_)), ada.cost_)
plt.xlabel('Iterations')
plt.ylabel('Cost')
Iteration: 30/30 | Cost 3.79 | Elapsed: 0:00:00 | ETA: 0:00:00
Text(0, 0.5, 'Cost')
Example 3 - Stochastic Gradient Descent
from mlxtend.data import iris_data
from mlxtend.plotting import plot_decision_regions
from mlxtend.classifier import Adaline
import matplotlib.pyplot as plt
# Loading Data
X, y = iris_data()
X = X[:, [0, 3]] # sepal length and petal width
X = X[0:100] # class 0 and class 1
y = y[0:100] # class 0 and class 1
# standardize
X[:,0] = (X[:,0] - X[:,0].mean()) / X[:,0].std()
X[:,1] = (X[:,1] - X[:,1].mean()) / X[:,1].std()
ada = Adaline(epochs=15,
eta=0.02,
minibatches=len(y), # for SGD learning
random_seed=1,
print_progress=3)
ada.fit(X, y)
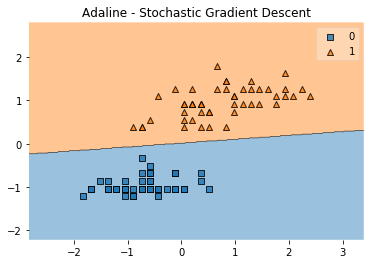
plot_decision_regions(X, y, clf=ada)
plt.title('Adaline - Stochastic Gradient Descent')
plt.show()
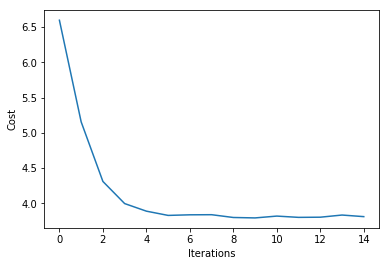
plt.plot(range(len(ada.cost_)), ada.cost_)
plt.xlabel('Iterations')
plt.ylabel('Cost')
plt.show()
Iteration: 15/15 | Cost 3.81 | Elapsed: 0:00:00 | ETA: 0:00:00
Example 4 - Stochastic Gradient Descent with Minibatches
from mlxtend.data import iris_data
from mlxtend.plotting import plot_decision_regions
from mlxtend.classifier import Adaline
import matplotlib.pyplot as plt
# Loading Data
X, y = iris_data()
X = X[:, [0, 3]] # sepal length and petal width
X = X[0:100] # class 0 and class 1
y = y[0:100] # class 0 and class 1
# standardize
X[:,0] = (X[:,0] - X[:,0].mean()) / X[:,0].std()
X[:,1] = (X[:,1] - X[:,1].mean()) / X[:,1].std()
ada = Adaline(epochs=15,
eta=0.02,
minibatches=5, # for SGD learning w. minibatch size 20
random_seed=1,
print_progress=3)
ada.fit(X, y)
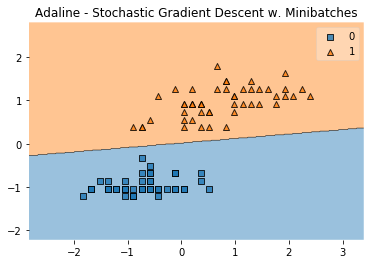
plot_decision_regions(X, y, clf=ada)
plt.title('Adaline - Stochastic Gradient Descent w. Minibatches')
plt.show()
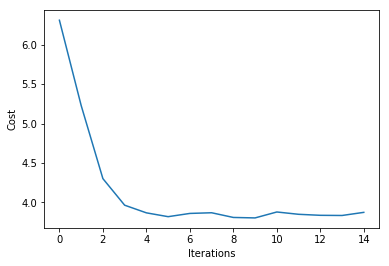
plt.plot(range(len(ada.cost_)), ada.cost_)
plt.xlabel('Iterations')
plt.ylabel('Cost')
plt.show()
Iteration: 15/15 | Cost 3.87 | Elapsed: 0:00:00 | ETA: 0:00:00
API
Adaline(eta=0.01, epochs=50, minibatches=None, random_seed=None, print_progress=0)
ADAptive LInear NEuron classifier.
Note that this implementation of Adaline expects binary class labels in {0, 1}.
Parameters
-
eta: float (default: 0.01)solver rate (between 0.0 and 1.0)
-
epochs: int (default: 50)Passes over the training dataset. Prior to each epoch, the dataset is shuffled if
minibatches > 1to prevent cycles in stochastic gradient descent. -
minibatches: int (default: None)The number of minibatches for gradient-based optimization. If None: Normal Equations (closed-form solution) If 1: Gradient Descent learning If len(y): Stochastic Gradient Descent (SGD) online learning If 1 < minibatches < len(y): SGD Minibatch learning
-
random_seed: int (default: None)Set random state for shuffling and initializing the weights.
-
print_progress: int (default: 0)Prints progress in fitting to stderr if not solver='normal equation' 0: No output 1: Epochs elapsed and cost 2: 1 plus time elapsed 3: 2 plus estimated time until completion
Attributes
-
w_: 2d-array, shape={n_features, 1}Model weights after fitting.
-
b_: 1d-array, shape={1,}Bias unit after fitting.
-
cost_: listSum of squared errors after each epoch.
Examples
For usage examples, please see https://rasbt.github.io/mlxtend/user_guide/classifier/Adaline/
Methods
fit(X, y, init_params=True)
Learn model from training data.
Parameters
-
X: {array-like, sparse matrix}, shape = [n_samples, n_features]Training vectors, where n_samples is the number of samples and n_features is the number of features.
-
y: array-like, shape = [n_samples]Target values.
-
init_params: bool (default: True)Re-initializes model parameters prior to fitting. Set False to continue training with weights from a previous model fitting.
Returns
self: object
get_params(deep=True)
Get parameters for this estimator.
Parameters
-
deep: boolean, optionalIf True, will return the parameters for this estimator and contained subobjects that are estimators.
Returns
-
params: mapping of string to anyParameter names mapped to their values.'
adapted from https://github.com/scikit-learn/scikit-learn/blob/master/sklearn/base.py Author: Gael Varoquaux gael.varoquaux@normalesup.org License: BSD 3 clause
predict(X)
Predict targets from X.
Parameters
-
X: {array-like, sparse matrix}, shape = [n_samples, n_features]Training vectors, where n_samples is the number of samples and n_features is the number of features.
Returns
-
target_values: array-like, shape = [n_samples]Predicted target values.
score(X, y)
Compute the prediction accuracy
Parameters
-
X: {array-like, sparse matrix}, shape = [n_samples, n_features]Training vectors, where n_samples is the number of samples and n_features is the number of features.
-
y: array-like, shape = [n_samples]Target values (true class labels).
Returns
-
acc: floatThe prediction accuracy as a float between 0.0 and 1.0 (perfect score).
set_params(params)
Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects
(such as pipelines). The latter have parameters of the form
<component>__<parameter> so that it's possible to update each
component of a nested object.
Returns
self
adapted from https://github.com/scikit-learn/scikit-learn/blob/master/sklearn/base.py Author: Gael Varoquaux gael.varoquaux@normalesup.org License: BSD 3 clause